TrANSCRIPTIONAL CONTROL OF AUTOPHAGY and LYSOSOMEs IN CANCER
Autophagy (also known as macroautophagy), is a cellular self-catabolic process, conserved from yeast to man, that mediates degradation of macromolecules and even whole organelles. Decreases in extracellular or intracellular nutrient levels serve as potent activators of autophagy while nutrient abundance suppresses this pathway. The process of autophagy involves generation of a double membrane vesicle (the autophagosome), which encapsulates cytoplasmic contents followed by delivery to lysosomes for cargo degradation. Substrates for autophagy-mediated degradation are diverse and include misfolded proteins, damaged organelles, protein aggregates and lipid droplets. The final step of of autophagy involves fusion with lysosomes - an acidic organelle in the cell responsible for breakdown of cargo. Deregulated autophagy and lysosome function has been implicated in the pathogenesis of cancer. We study a particularly aggressive cancer - pancreatic ductal adenocarcinoma (PDA) - which display constitutive autophagy activation and increased lysosome biogenesis.
We have identified the MiT/TFE family of transcription factors (TFE3, MITF, and TFEB) as master regulators of autophagy-lysosome activation in PDA. Constitutive localization of these factors in the nucleus facilitates transcriptional control of the biogenesis and function of the autophagosome-lysosome system. We are interested in understanding the regulatory circuits controlling sustained autophagy and lysosome induction both at the transcriptional and signaling levels in PDA cells.
ORGANELLE FUNCTION in Cancer
How do cancer cells exploit enhanced lysosome activity to gain a survival and growth advantage? To answer this question we must first understand the qualitative differences between lysosomes in cancer cells and normal cells. To do so, we use a combination of affinity based organelle purification, proteomics and functional assays in cell and tumor based model systems to build a map of the cancer cell lysosome. Through this analysis we will also determine how changes in lysosome composition and function contribute to tumor evolution, progression and spread.
Mechanisms of tumor heterogeneity and cellular plasticity
Activating mutations in KRAS are present in >90% of PDA tumors. Despite this common driver mutation, PDA tumors display considerable heterogeneity. We are interested in determining how cellular plasticity mediated by dynamic changes in the activation status of embryonic developmental programs, stress response pathways and metabolic reprogramming enables tumor cell adaptation to a variety of external and internal cellular stresses during primary tumor growth and metastasis.
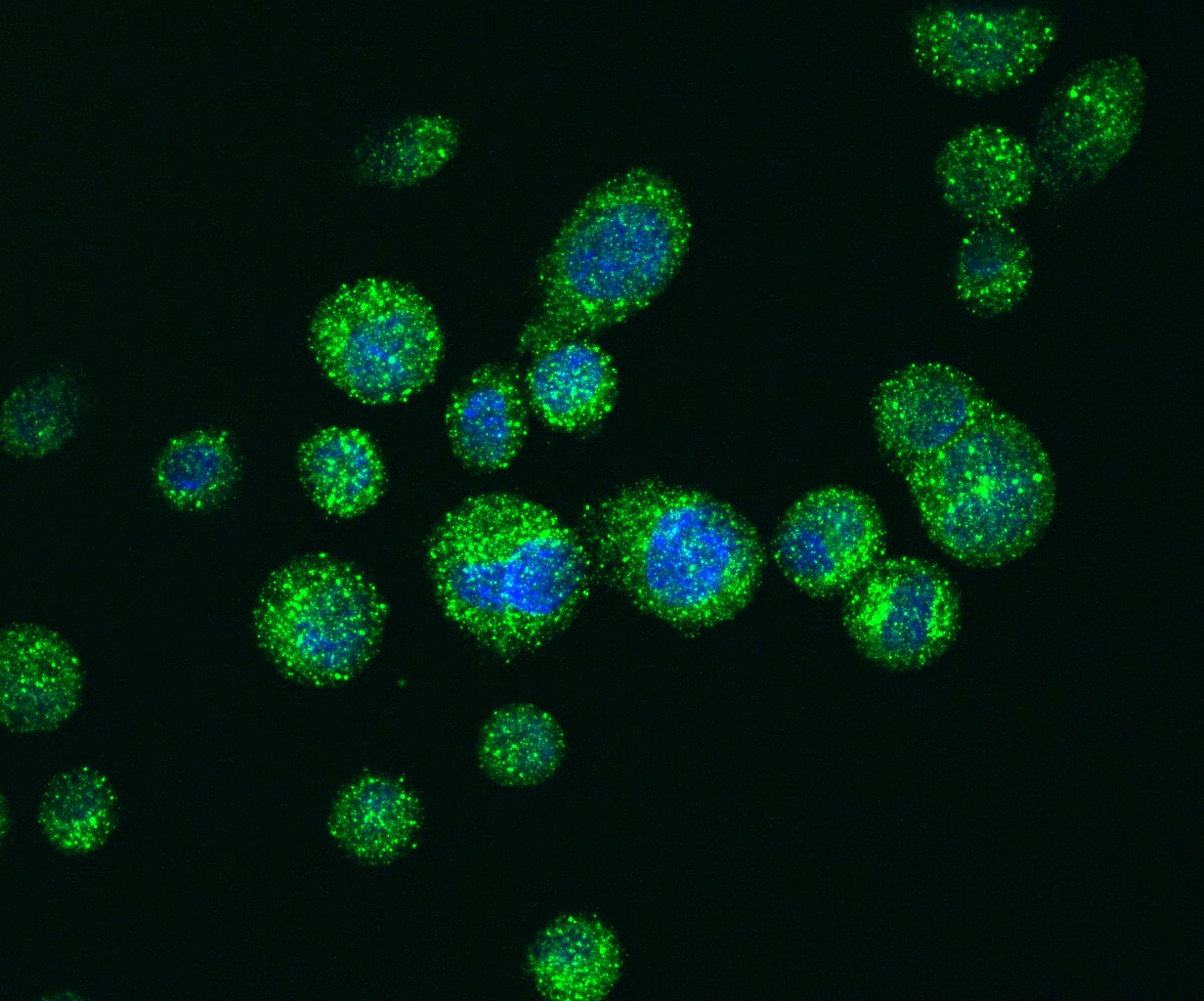
Immuno-fluorescence staining of LC3 positive autophagosomes (green) in PDA cells.
Heatmap showing coordinate up-regulation of autophagy-lysosome genes in PDA versus matched normal tissue.
Immuno-fluorescence staining of lysosomes in PDAC tumors.